My first postdoctoral publication is now online! In this excellent collaboration between @WeitzmanLab, @DanDepledge, @TheHornerLab, and @mason_lab we used @nanopore #directRNA sequencing to reveal how m6A RNA modifications mediate Adenoviral RNA splicing. https://doi.org/10.1038/s41467-020-19787-6
A thread(2/9): In 1976 it was shown that Adenovirus (AdV) contained m6A RNA modifications within mature viral RNA transcripts, but no biological function of this mark was able to be assigned using the technology of the era.
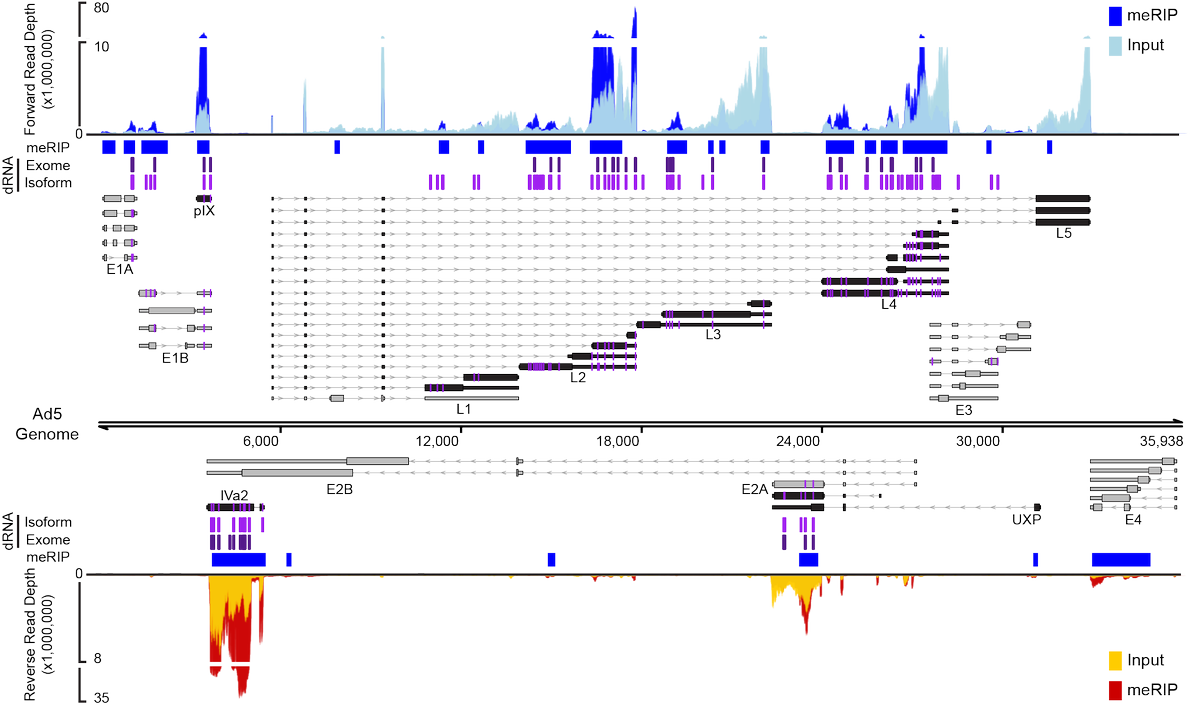
(3/9) Exactly 40 years later we performed meRIP-seq during AdV infection and quickly discovered that typical short RNA reads were going to be insufficient to accurately map RNA modifications onto the highly spliced and overlapping transcriptome of this DNA virus.
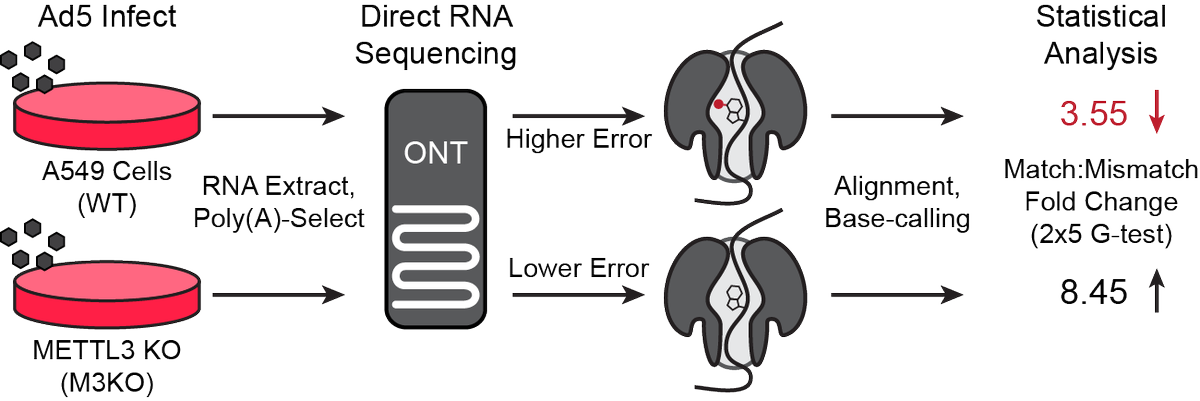
(4/9) What we needed was a tool that could phase a single modification while containing information about upstream and downstream splicing events. Swooping in to our rescue came @DanDepledge, armed with a @nanopore minION and a very interesting idea.
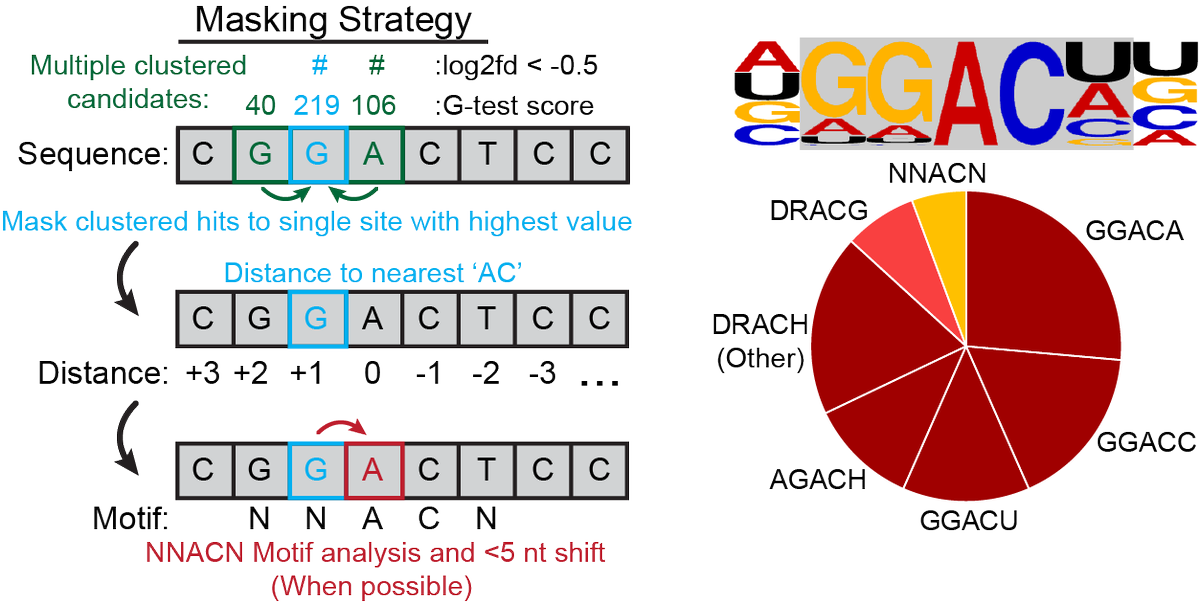
(5/9) RNA mods cause sequencing errors when transiting a pore. By comparing viral RNA generated in wildtype cells to METTL3 KO cells we were able to use a statistical pipeline (DRUMMER) to call sites of METTL3-dependent m6A additions at the single-nucleotide level.
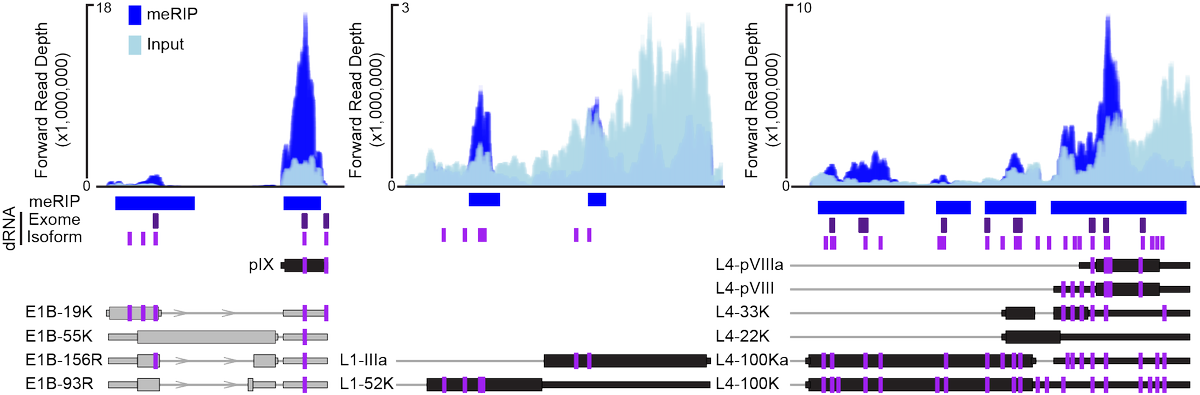
(6/9) Furthermore, by throwing out all ambiguously mapped RNA reads we were able to increase our signal to noise ratio and find even more modifications that could now be mapped to individual transcript isoforms!
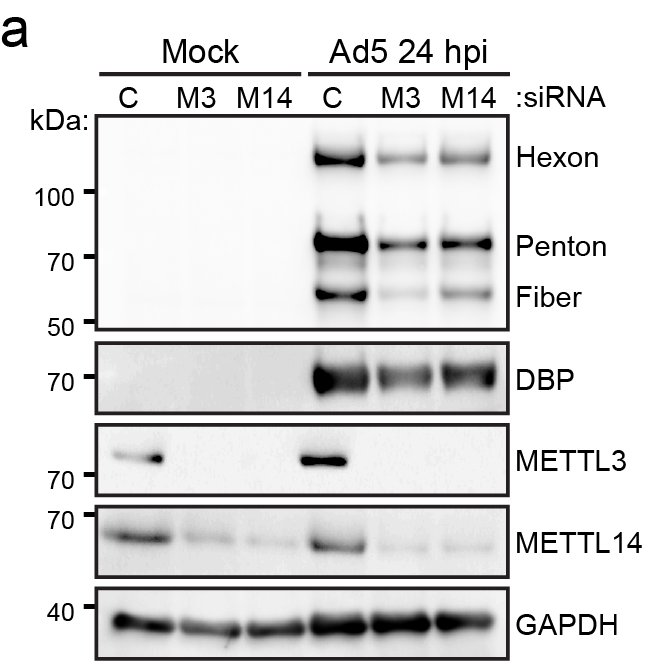
(7/9) While the majority of viral early and late transcripts were modified, we only noticed a significant decrease in late stage viral RNAs or proteins after loss of METTL3. Ultimately, it was shown that m6A was necessary for splicing of late transcripts with long introns.
(8/9) Overall, this exciting collaboration used many new technologies to answer an old question. In particular, the use of long-read sequencing allowed for the re-annotation of the AdV transcriptome yielding surprising results I hope to tell you all about very soon.
(9/9) For now I will leave you with a bioRxiv preview: https://www.biorxiv.org/content/10.1101/2019.12.13.876037v1

 Read on Twitter
Read on Twitter