Beyond stoked to have published the first paper out of the Chanda Lab and my first first-author paper in #JNeurosci! It's been truly exciting unpacking the mechanism of this uncharacterized Autism-linked mutation @SfNJournals 1/N https://www.jneurosci.org/content/early/2020/11/24/JNEUROSCI.0404-20.2020
2/ This is essentially a case study of an uncharacterized NLGN4 mutation in a kid diagnosed with severe Autism. We found that the mutation strongly impairs NLGN4 trafficking in human neurons, increasing EPSC amplitude and frequency likely by inhibiting GluA1 synaptic localization
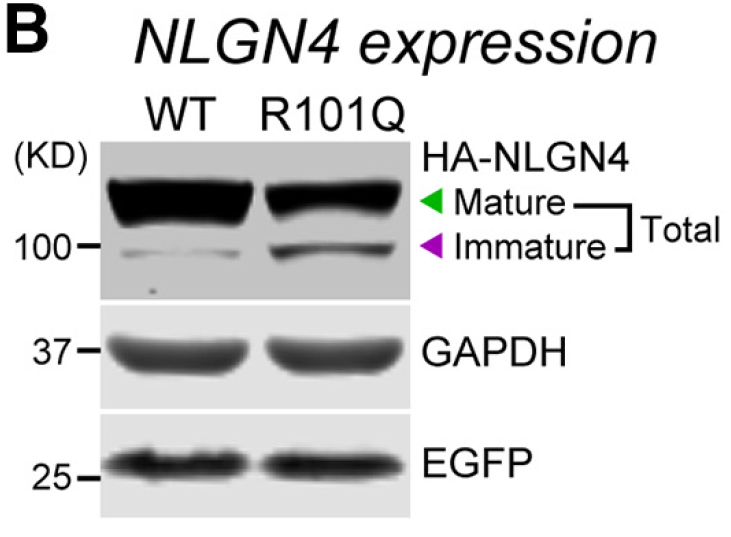
3/ The mutation (R101Q) doesn't reduce NLGN4 expression that much, but affects its glycosylation. This in turn causes the mutant protein to get stuck in the secretory P/W, specifically the ER and Golgi
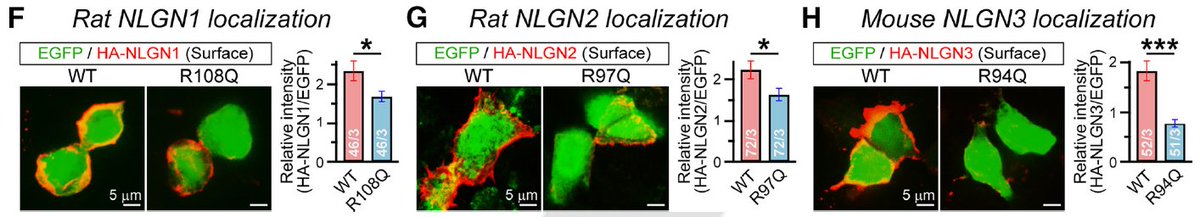
4/ What's interesting is the mutation site, R101, is right next to an N-linked glycosylation sequon (N102). An N102A mutation phenocopies R101Q and these two residues are conserved in NLGN1-3. Equivalent R-Q mutations in NL1-3 have similar effects
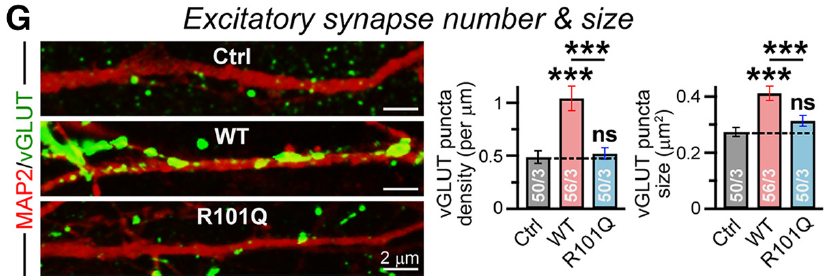
5/ We whipped up human neurons using two protocols, NSC-derived and direct differentiation from ESCs. In both, WT NL4 overexpression increases vGlut and Synapsin density, but the mutant doesn't at all
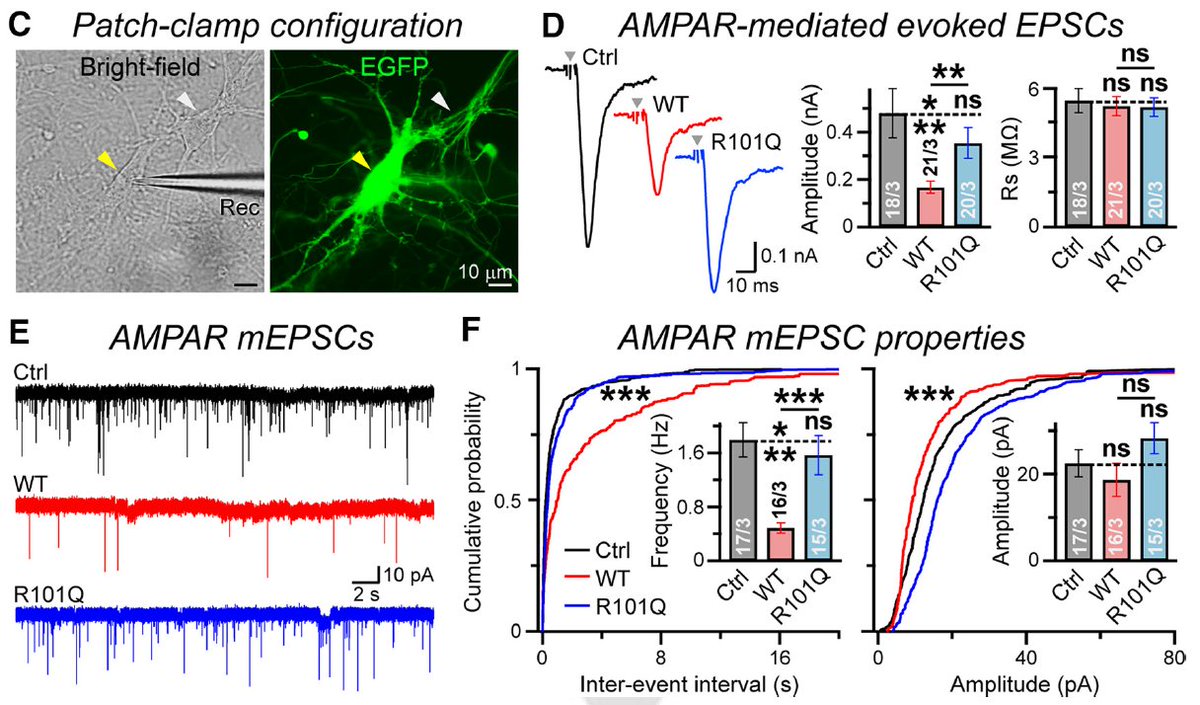
6/ Confusingly, NL4-R101Q actually increases excitatory currents vs WT, for both evoked and mini EPSCs.
7/ As a note, the vGlut and Synapsin density increase is probably an overexpression artifact. NLGN KO studies don't reveal any change in synapse number, but overexpression tends to stabilize transient synapses. Point is, R101Q isn't doing what WT normally does
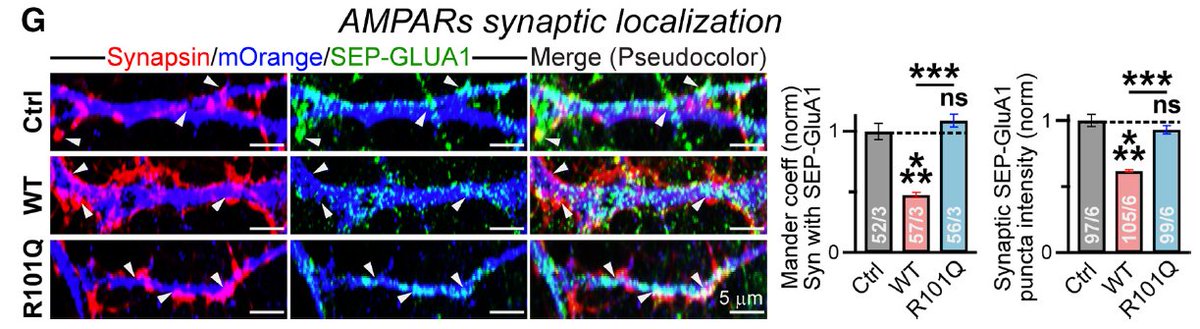
8/ Hinting at NLGN4's true role, we see that while WT overexpression increases Synapsin levels, GluA1 is pushed out of the synapse (a possible mechanism to explain the EPSC effects)

 Read on Twitter
Read on Twitter