In 2020 with @DrOlaLandgren we revealed how Melphalan has strong mutagenic activity on myeloma cells. This can understandably create fear for patients who have undergone transplant, and we have received many questions from them. Here a thread on what we know so far #msmm 





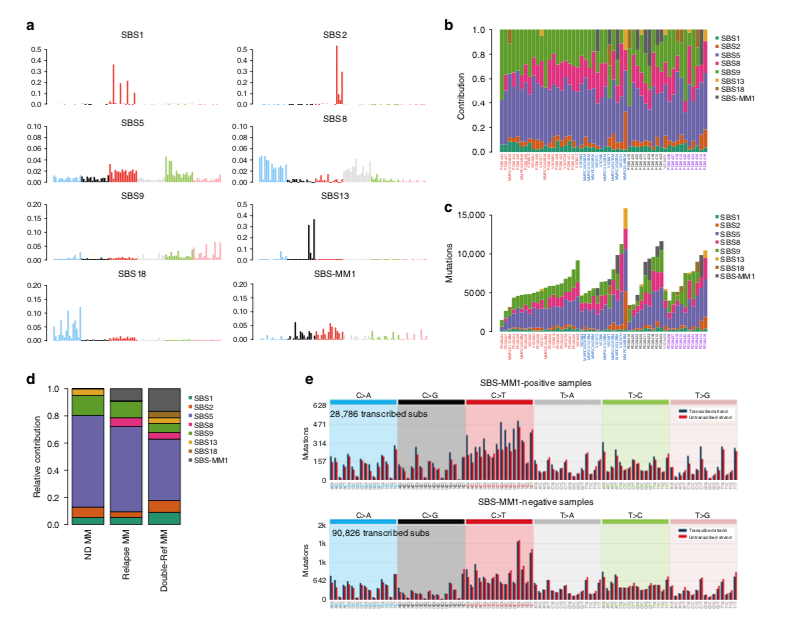
Using whole-exome and genome sequencing data, we can capture the majority of mutations in cancer. Using bioinformatic algorithms we can identify and quantify which mutational processes are involved and responsible for this catalog of mutations (i.e. mutational signatures)
In 2017 Weinhold and @DrGarethMorgan1 reported a new mutational signature in relapsed multiple myeloma https://doi.org/10.1182/blood-2016-06-723007. In 2018-2019 with @nickbolli we found the same signature (MM1; https://rdcu.be/ccTQl and https://rdcu.be/ccTQm ), still without knowing its etiology
This year, with @EHRustad @DrOlaLandgren we linked this distinct signature (named SBS-MM1) to Melphalan: https://rdcu.be/ccTQB . This association was validated on external data (CoMMpass @theMMRF ) and on cell lines
@BachisioZic and @nickbolli further confirmed the enrichment of this signature among relapsed and refractory multiple myeloma patients: 10.1182/bloodadvances.2019000779
Mid 2020, we showed that myeloma cells exposed to Melphalan always acquired these mutations, detectable in bulk sequencing only if one cell takes the clonal dominance and expands. This is why we don't see SBS-MM1 in all post-melphalan cases https://rdcu.be/ccVuO
This concept of “single cell expansion” and the utility of chemotherapy-barcode in timing cancer evolution has been also very well described by @oriol_pich and @nlbigas: https://rdcu.be/ccTQR and https://www.biorxiv.org/content/10.1101/237941v1.
Not all patients acquired these mutations! A fraction of patients might relapse through the progression of myeloma cells infused back into the body during transplant. These cells have never been exposed to melphalan and therefore they will not have any melphalan-related mutations
At the end of 2020, two ASH abstracts from Munshi’s and Weinhold’s labs independently confirmed SBS-MM1 as the melphalan mutational signature in relapsed multiple myeloma #ASH20: https://ash.confex.com/ash/2020/webprogram/Paper139014.html and https://ash.confex.com/ash/2020/webprogram/Paper141611.html
While melphalan mutations contribute to up to 10-20% of nonsynonymous mutations at relapse, their role in tumor progression is unclear. It is possible that these mutations might introduce new driver mutations making myeloma more aggressive, but this is likely a small contribution
Apart from tumor, after transplant, all replicating tissues in our body will acquire Melphalan-related mutations, and this might partially explain the increased risk of therapy-related malignancies in myeloma patients after transplant.
These data represent a biological explanation of what has been known for decades. Melphalan is effective in myeloma, but also toxic. Future new therapies and MRD-oriented strategies will tell if this approach can be reserved for a select group of patients, rather than all

 Read on Twitter
Read on Twitter